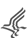| Track | Description | Hydra AEP | Hydra oligactis |
| ATAC_Merged | Depth normalized read count density for AEP ATAC-seq data, combines all three biological replicates |  | |
| ATAC_Rep1 | Depth normalized read count density for AEP ATAC-seq data biological replicate 1 |  | |
| ATAC_Rep2 | Depth normalized read count density for AEP ATAC-seq data biological replicate 2 |  | |
| ATAC_Rep3 | Depth normalized read count density for AEP ATAC-seq data biological replicate 3 |  | |
| ATAC_actin:gfp_neural_Merged | Depth normalized read count density for neuron enriched ATAC-seq data collected from Tg(actin1:GFP)rs3-in transgenic animals, combines all four biological replicates |  | |
| ATAC_actin:gfp_neural_Rep1 | Depth normalized read count density for neuron enriched ATAC-seq data collected from Tg(actin1:GFP)rs3-in transgenic animals, biological replicate 1 |  | |
| ATAC_actin:gfp_neural_Rep2 | Depth normalized read count density for neuron enriched ATAC-seq data collected from Tg(actin1:GFP)rs3-in transgenic animals, biological replicate 2 |  | |
| ATAC_actin:gfp_neural_Rep3 | Depth normalized read count density for neuron enriched ATAC-seq data collected from Tg(actin1:GFP)rs3-in transgenic animals, biological replicate 3 |  | |
| ATAC_actin:gfp_neural_Rep4 | Depth normalized read count density for neuron enriched ATAC-seq data collected from Tg(actin1:GFP)rs3-in transgenic animals, biological replicate 4 |  | |
| ATAC_tba1c:mNeonGreen_neural_Merged | Depth normalized read count density for neuron enriched ATAC-seq data collected from Tg(tba1c:mNeonGreen)cj1-gt transgenic animals, combines all two biological replicates |  | |
| ATAC_tba1c:mNeonGreen_neural_Rep1 | Depth normalized read count density for neuron enriched ATAC-seq data collected from Tg(tba1c:mNeonGreen)cj1-gt transgenic animals, biological replicate 1 |  | |
| ATAC_tba1c:mNeonGreen_neural_Rep2 | Depth normalized read count density for neuron enriched ATAC-seq data collected from Tg(tba1c:mNeonGreen)cj1-gt transgenic animals, biological replicate 2 |  | |
| H3K27me3_Merged_Reps | Depth normalized read count density for AEP cut&tag targeting H3K27me3, combines all three biological replicates |  | |
| H3K27me3_Rep1 | Depth normalized read count density for AEP cut&tag targeting H3K27me3, biological replicate 1 |  | |
| H3K27me3_Rep2 | Depth normalized read count density for AEP cut&tag targeting H3K27me3, biological replicate 2 |  | |
| H3K27me3_Rep3 | Depth normalized read count density for AEP cut&tag targeting H3K27me3, biological replicate 3 |  | |
| H3K4me1_Merged_Reps | Depth normalized read count density for AEP cut&tag targeting H3K4me1, combines all three biological replicates |  | |
| H3K4me1_Rep1 | Depth normalized read count density for AEP cut&tag targeting H3K4me1, biological replicate 1 |  | |
| H3K4me1_Rep2 | Depth normalized read count density for AEP cut&tag targeting H3K4me1, biological replicate 2 |  | |
| H3K4me1_Rep3 | Depth normalized read count density for AEP cut&tag targeting H3K4me1, biological replicate 3 |  | |
| H3K4me3_Merged_Reps | Depth normalized read count density for AEP cut&tag targeting H3K4me3, combines all three biological replicates |  | |
| H3K4me3_Rep1 | Depth normalized read count density for AEP cut&tag targeting H3K4me3, biological replicate 1 |  | |
| H3K4me3_Rep2 | Depth normalized read count density for AEP cut&tag targeting H3K4me3, biological replicate 2 |  | |
| H3K4me3_Rep3 | Depth normalized read count density for AEP cut&tag targeting H3K4me3, biological replicate 3 |  | |
| IgG_Merged_Reps | Depth normalized read count density for control AEP cut&tag using non-specific antibodies, combines all three biological replicates |  | |
| IgG_Rep1 | Depth normalized read count density for control AEP cut&tag using non-specific antibodies, biological replicate 1 |  | |
| IgG_Rep2 | Depth normalized read count density for control AEP cut&tag using non-specific antibodies, biological replicate 2 |  | |
| IgG_Rep3 | Depth normalized read count density for control AEP cut&tag using non-specific antibodies, biological replicate 3 |  | |
| Chemi_conservation | 100bp moving window conservation score for the pairwise alignment between C. hemisphaerica and H. vulgaris strain AEP (score range: 0-1) |  | |
| HOLI1_conservation | 100bp moving window conservation score for the pairwise alignment between H. oligactis and H. vulgaris strain AEP (score range: 0-1) |  | |
| Hv105_conservation | 100bp moving window conservation score for the pairwise alignment between H. vulgaris strain 105 and H. vulgaris/ strain AEP (score range: 0-1) |  | |
| Hvirid_conservation | 100bp moving window conservation score for the pairwise alignment between H. viridissima and H. vulgaris strain AEP (score range: 0-1) |  | |
| all4_conservation | 100bp moving window conservation score for all 4 non-AEP genomes in the alignment (score range: 0-4) |  | |
| HVAEP Gene Models | Hydra AEP assembly gene models |  | |
| InterProScan | Interpro results for the longest protein isoforms from the AEP protein models |  | |
| BLASTP_NR | Top 5 hits for the longest AEP protein isoforms blasted against the NCBI nr protein db. In standard tabular format with the addition of a column for the corresponding definition field for each hit's accession number |  | |
| BLASTP_UniProt | Top 5 hits for the longest AEP protein isoforms blasted against the reviewed swissprot protein db. In standard tabular format |  | |
| HVAEP_repeats | Hydra AEP predicted repetitive regions, generated by RepeatMasker using repeat families from Dfam (eumetazoa db) and Repeatmodeler2 |  | |
| consensus_ATAC_peaks | AEP genome coordinates for biologically reproducible ATAC-seq peaks |  | |
| consensus_H3K27me3_peaks | AEP genome coordinates for biologically reproducible H3K27me3 cut&tag peaks |  | |
| consensus_H3K4me1_peaks | Hydra AEP genome coordinates for biologically reproducible H3K4me1 cut&tag peaks |  | |
| consensus_H3K4me3_peaks | Hydra AEP genome coordinates for biologically reproducible H3K4me3 cut&tag peaks |  | |
| conserved_ATAC_peaks | Hydra AEP genome coordinates for evolutionarily conserved ATAC-seq peaks (subset of HVAEP1_consensus_ATAC_peaks.bed) |  | |
| conserved_H3K27me3_peaks | Hydra AEP genome coordinates for evolutionarily conserved H3K27me3 Cut&Tag peaks |  | |
| conserved_H3K4me1_peaks | Hydra AEP genome coordinates for evolutionarily conserved H3K4me1 Cut&Tag peaks |  | |
| conserved_H3K4me3_peaks | Hydra AEP genome coordinates for evolutionarily conserved H3K4me3 Cut&Tag peaks |  | |
| conserved_TFBS | Hydra AEP genome coordinates for evolutionarily conserved putative transcription factor binding sites |  | |
| hic_chromatin_domains | Hydra AEP genome coordinates for chromatin contact domains. Predicted using Hi-C data with a 16 Kb bin size |  | |
| MASK | Predicted repeats in the Hydra AEP and H. oligactis genomes, generated by repeatmasker using repeat families from Dfam (eumetazoa db) and repeatmodeler2 |  |  |
| SCF | Assembled genomic scaffolds (SCF) appear as solid black tracks with intermittent gaps shaded bright pink |  |  |
| Reference sequence | The Hydra AEP and Hydra oligactis genomic sequences and corresponding six-frame translations depicted when fully zoomed-in |  |  |
| HOLI Gene Models | Hydra oligactis gene models generated by braker2 | |  |
| HOLI Predicted Transcripts | Hydra oligactis predicted transcripts generated by braker2 | |  |